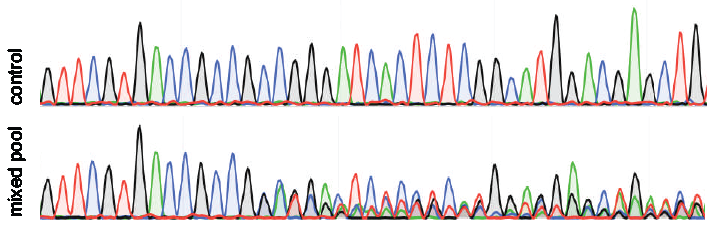
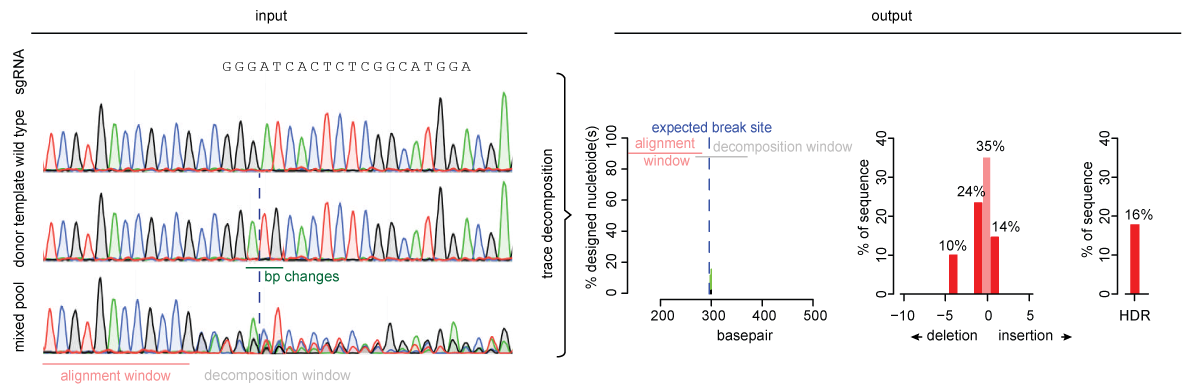
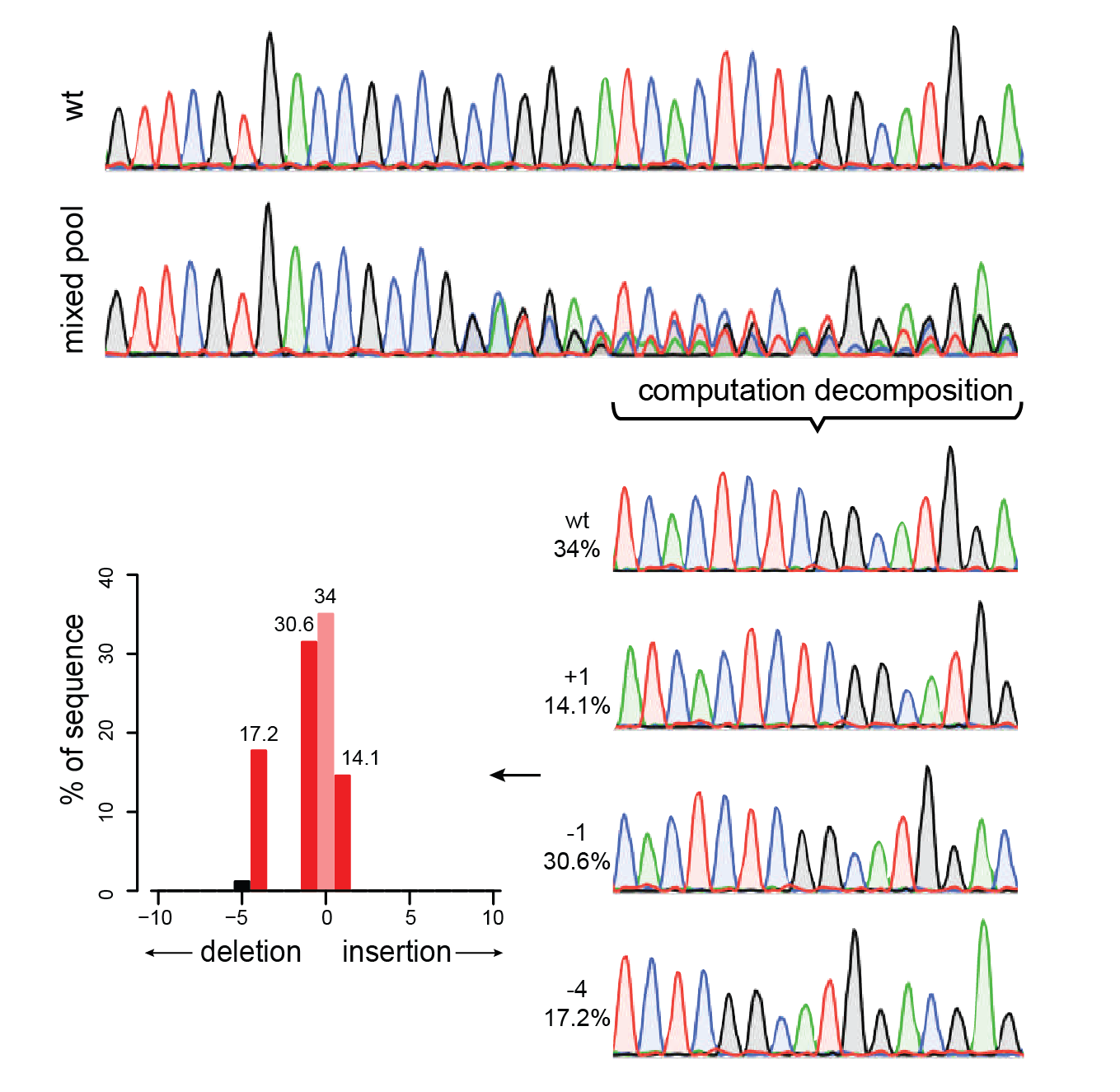
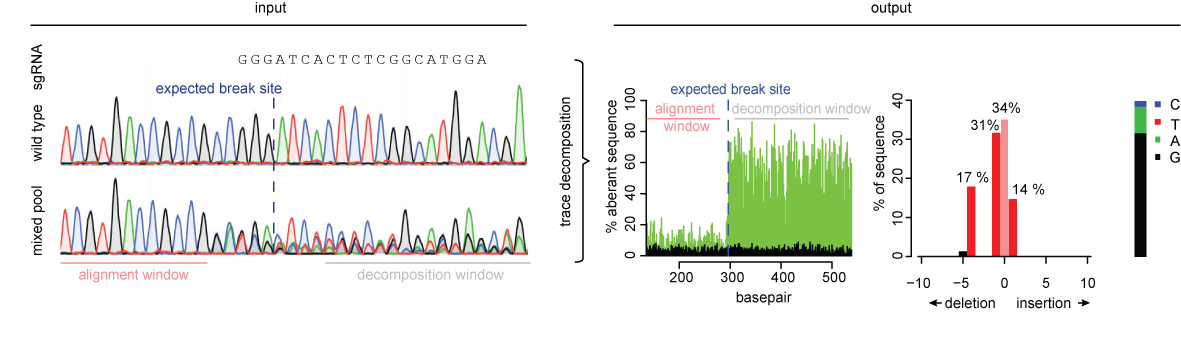
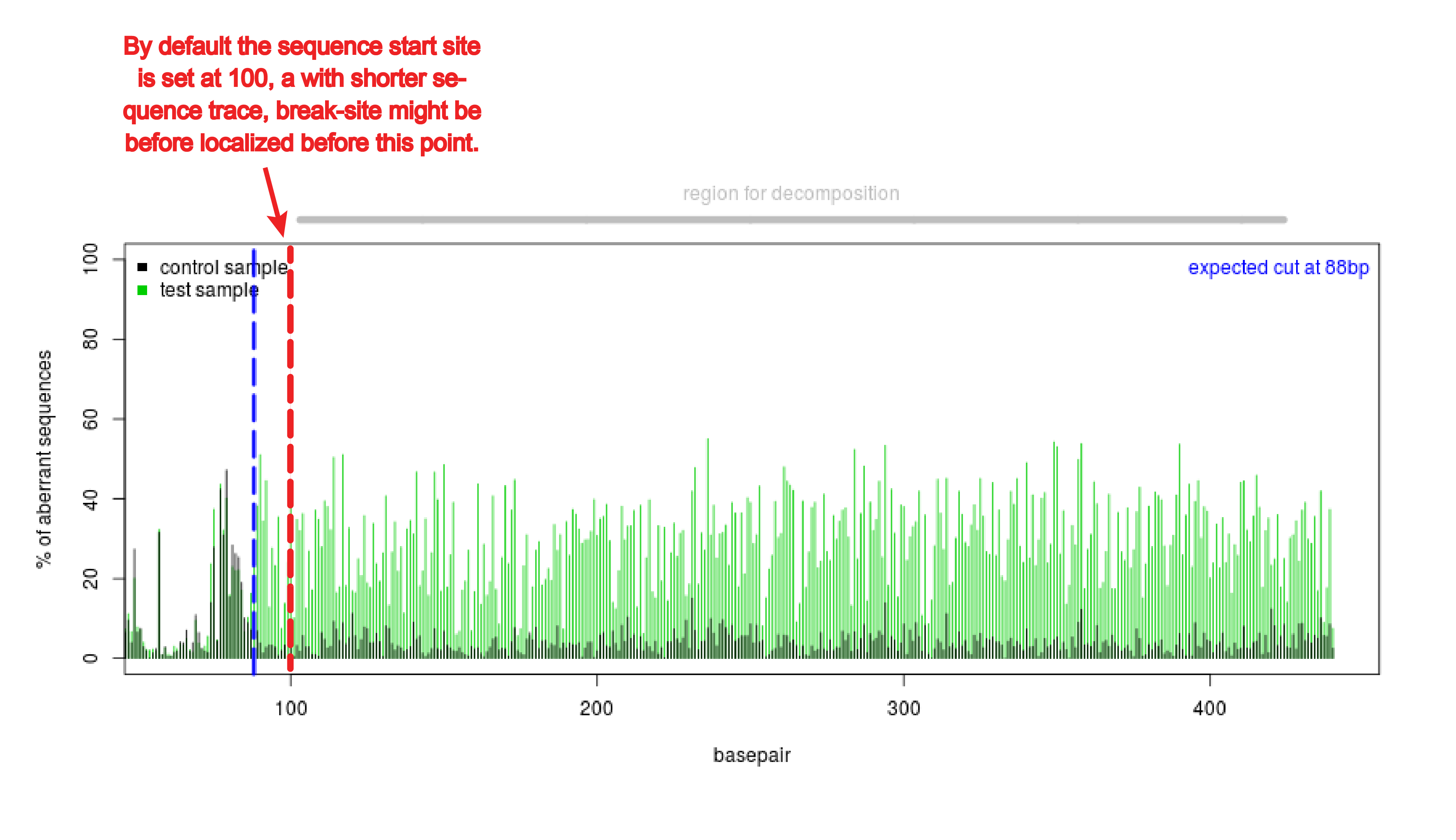
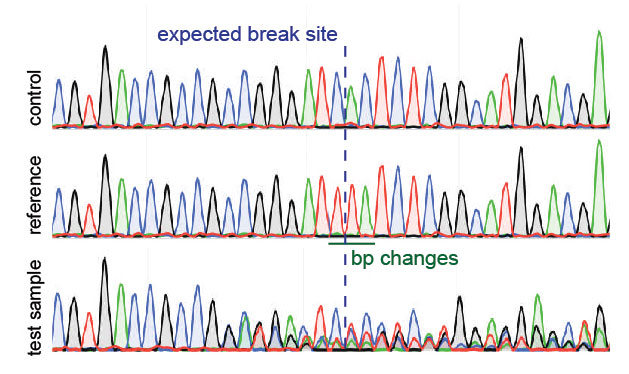
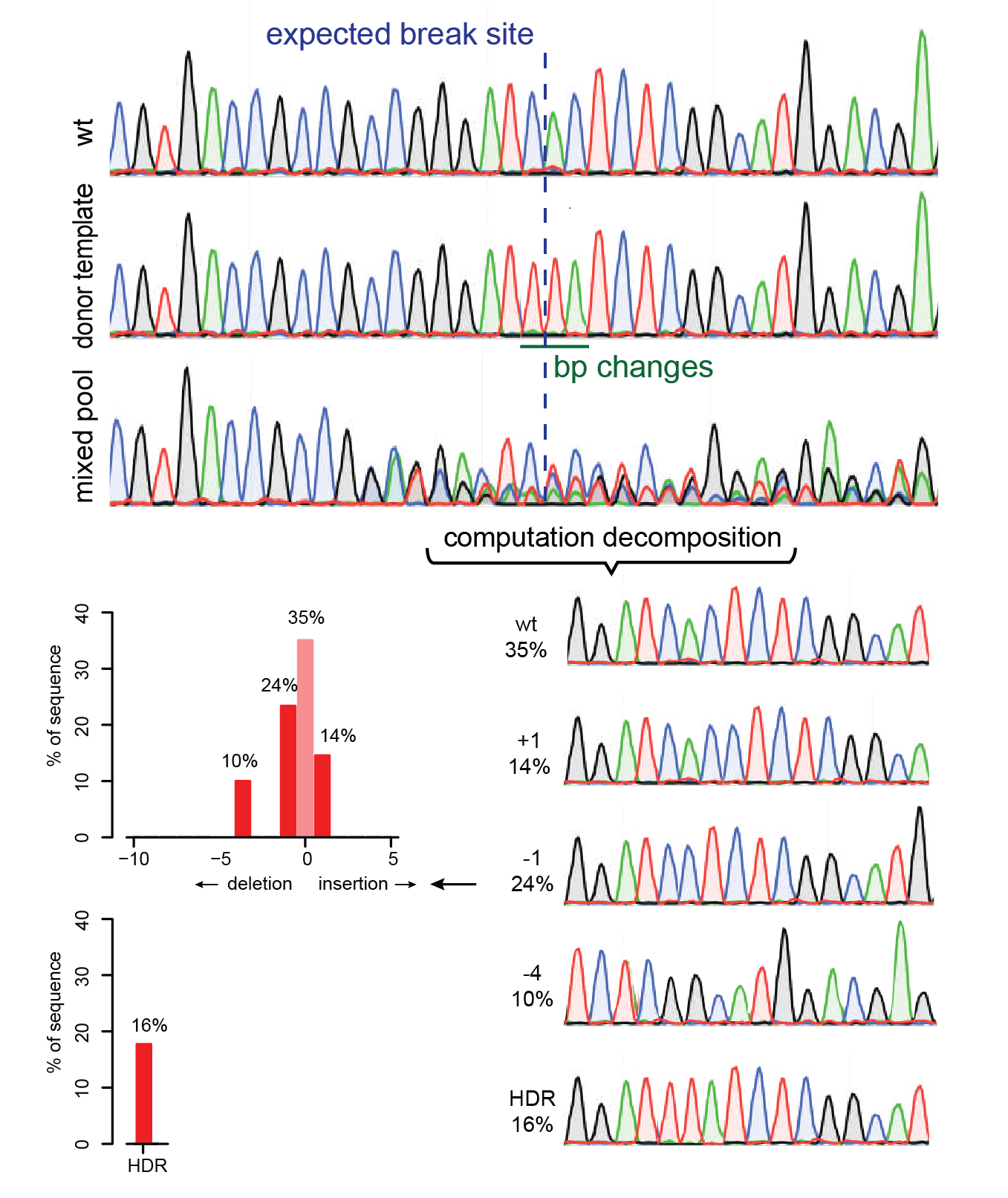
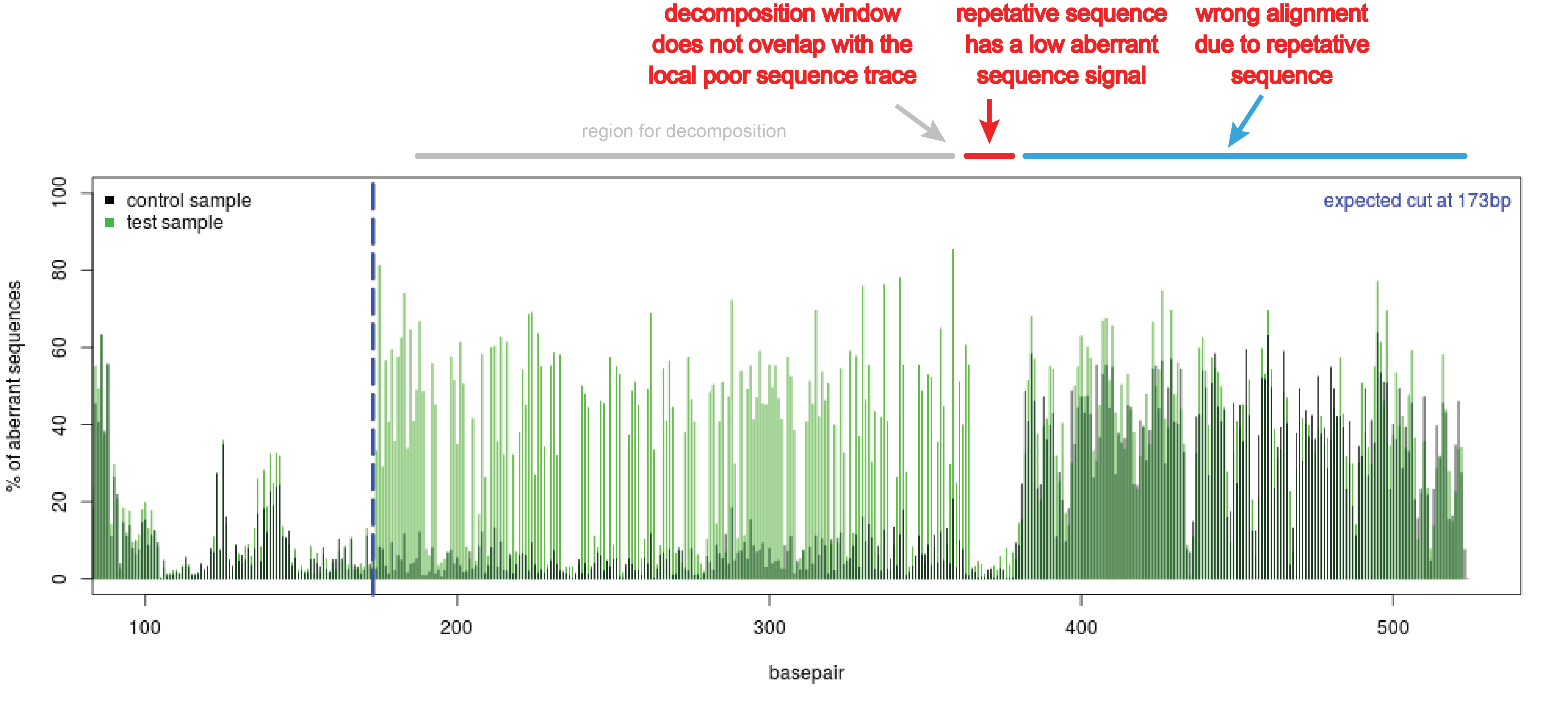
TIDE sequencing analysis of light-mediated indel mutation HEK293T cells... | Download Scientific Diagram

Netherlands Cancer Institute Grants Exclusive License to Desktop Genetics for TIDE | Technology Networks
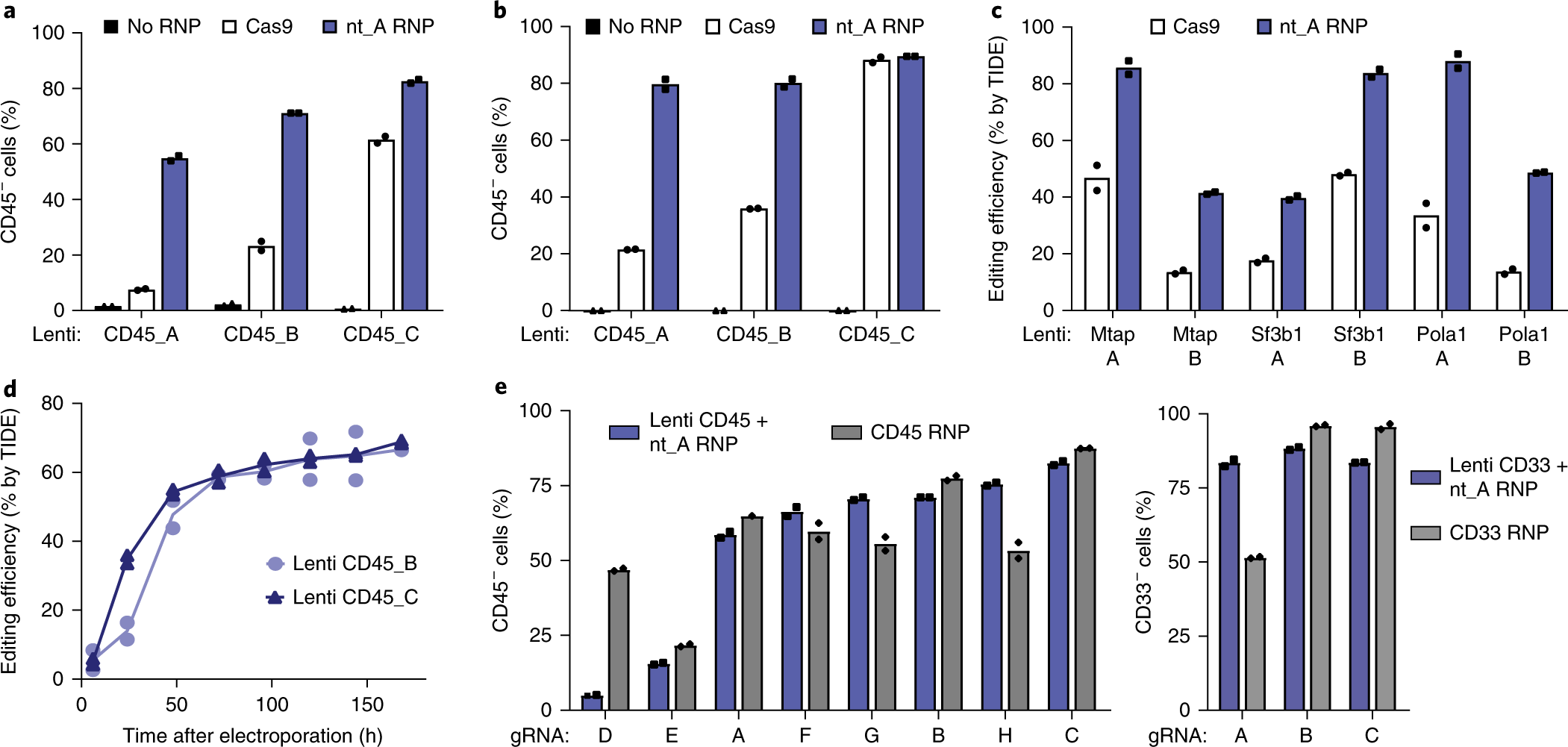
Guide Swap enables genome-scale pooled CRISPR–Cas9 screening in human primary cells | Nature Methods

Increasing CRISPR Efficiency and Measuring Its Specificity in HSPCs Using a Clinically Relevant System: Molecular Therapy - Methods & Clinical Development

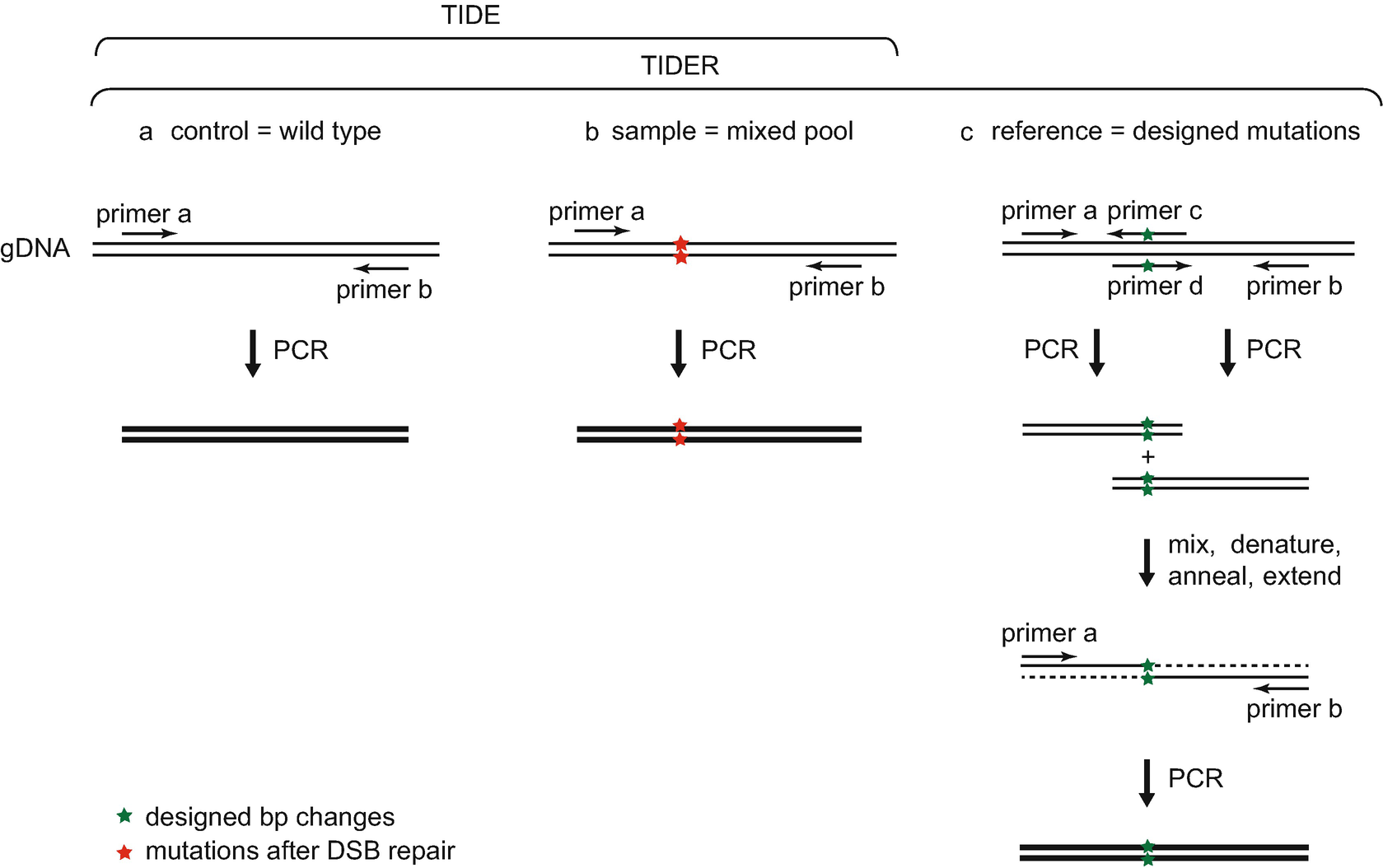
Analysis of gene modification conditions using TIDE software. The bar... | Download Scientific Diagram
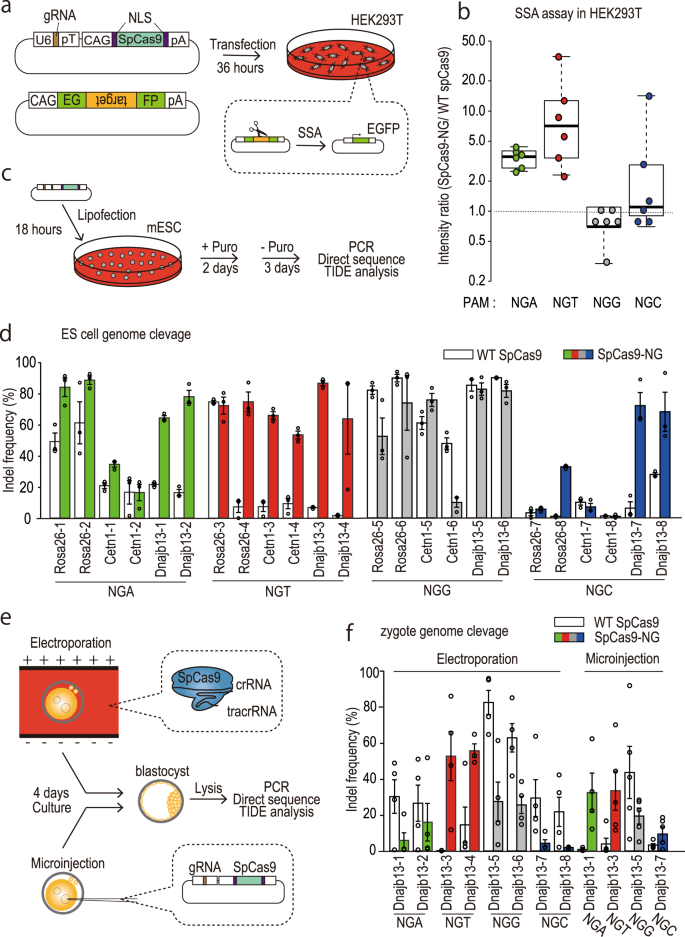
Precise CAG repeat contraction in a Huntington's Disease mouse model is enabled by gene editing with SpCas9-NG | Communications Biology
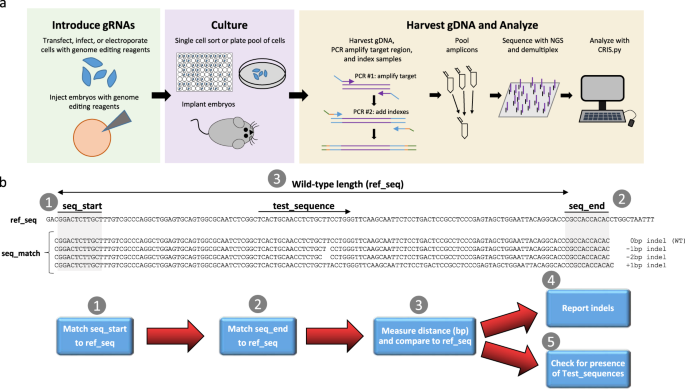
CRIS.py: A Versatile and High-throughput Analysis Program for CRISPR-based Genome Editing | Scientific Reports